Last modified 2025-10-23 |
Use the Read Samples SDK Method (Tutorial)
| Abbreviations Key | |||||
df | DataFrame | pd | pandas | ||
guid | globally unique identifier | QC | quality control | ||
| HISE | Human Immune System Explorer | SDK | software development kit | ||
hp | hisepy | SLIMS | simplified lab information management system | ||
| IDE | integrated development environment | UUID | universally unique identifier |
At a Glance
This document explains how to use read_samples() to retrieve sample metadata in HISE. This method lets you read or search the SampleStatus materialized view, enabling quick retrieval of sample metadata from complex queries or aggregations that would otherwise be slow to compute on demand. The read_samples() method bridges the workflow between lab data in SLIMS and analytical data in HISE.
Method signature
 read_samples()
read_samples()
hp.read_samples(
sample_ids: list = None,
query_dict: dict = None,
to_df=True,
)
 readSamples()
readSamples()
readSamples( sampleIds = NULL, query = NULL, toDF = TRUE, cleanLabCols = FALSE )
Parameters
The parameters for this method are listed in the following table. In each key:value pair, the value must be of type list.
Instructions
 Import libraries
Import libraries
To get started, set up your environment to interact with HISE programmatically and access all available SDK functions. For details, see Use Hise SDK Methods.
1. Navigate to HISE, and use your organizational email address to sign in.
2. Open an IDE. For instructions, see Create Your First HISE IDE (Tutorial).
3. For programmatic access to HISE functions and efficient handling of tabular data, import the Python SDK and the pandas library.
# Import hisepy and pandasimport hisepy as hp
import pandas as pd
 Define sample IDs
Define sample IDs
In this step, you define the sample IDs for this notebook. For details, see Use Advanced Search (Tutorial).
1. Retrieve your own set of study-specific sample IDs from SLIMS or an advanced search, and then define them as shown below. (This example uses specimenGuids as sample IDs.)
# Define the sample IDs used in this analysis SAMPLEIDS = ['4524bbe0-dcc7-47c9-8823-5a395efb83ec', '488828d9-1944-43e8-b5d0-a29b5816ab53', 'b89e4102-73dd-4e6f-8f25-cddaf9d2468d']

 Return dictionary output
Return dictionary output
When you pass in your sample IDs, a dictionary with four keys is returned. Each key is structured as a pandas DataFrame containing detailed information.
1. Pass your list of sample IDs to read_samples().
# Return the dictionary output
sample_out = hp.read_samples(sample_ids=SAMPLEIDS)
sample_out.keys()
The following dictionary keys are returned.
dict_keys(['metadata', 'specimens', 'survey', 'labResults']) |

| To see what's in a given dictionary key, use the following format:
specimens key:
|
These keys contain the metadata shown in the following table.
| Key | Description |
|---|---|
metadata | Dates and times of sample collection |
| Paths to SLIMS storage locations | |
| QC flags | |
specimens | Percentages of various cell types |
| Viability metrics | |
| Aliquot tracking through SLIMS container IDs | |
survey | Self-reported patient outcomes |
| Symptoms temporally aligned with data collection events | |
| Medication compliance | |
labResults | Cytokine levels |
| Flow cytometry gating strategies | |
| SLIMS-generated analyses |
 Merge DataFrames
Merge DataFrames
When working with sample data, you often have multiple tables (called DataFrames) containing various types of information about the same set of samples. Each table is identified by a unique key, such as sampleKitGuid or kitId. Sometimes it's helpful to merge DataFrames—that is, to combine information from different tables into a single table—so you can analyze or visualize all relevant data together. For example, you might want to combine metadata and lab results, filter samples based on specific criteria, or correlate lab data with a particular survey response. The following merge is just one example.
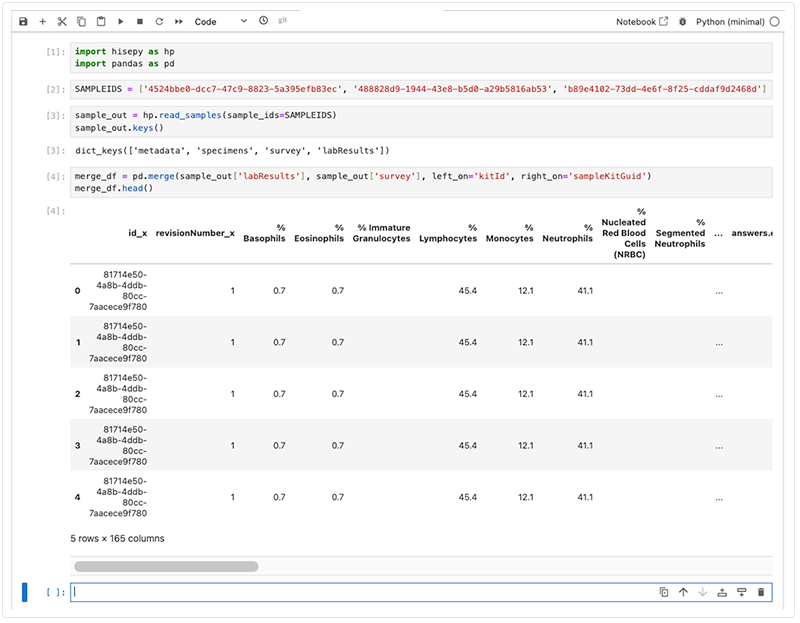
1. To place lab results and survey data into a single row for easier downstream analysis, enter the following code into a new cell in your IDE.
# Merge lab results with survey data
merge_df = pd.merge(sample_out['labResults'], sample_out['survey'], left_on='kitId', right_on='sampleKitGuid')
merge_df.head()

 Related Resources
Related Resources