Schedule and Troubleshoot Notebook Jobs (Tutorial)
| Abbreviations Key | |
| h-sne | hierarchical stochastic neighbor embedding |
| HISE | Human Immune System Explorer |
| IDE | integrated development environment |
| sf | simple features (for R) |
At a Glance
This article offers tips, tricks, and troubleshooting advice for executing code in a Jupyter notebook environment. It also explains how to schedule computationally intensive notebook tasks.
Navigate Your Notebook
To navigate Jupyter notebooks, try the following tips:
- Use Shift+Enter to run the code in the current cell.
- Press Tab to autocomplete code.
- To set up packages or environments when you boot up, use the startup script in the home directory. Be aware that if this startup script fails, it can delay the booting of your instance.
- Rather than use a Jupyter notebook to execute shell commands, you can use an exclamation point (
!) at the beginning of your command in Python, or asystem(‘COMMAND’)in R. - To see the execution time of a cell, use
%%timein Python, or something likestart <- Sys.time(); y <- 1:5; end <- Sys.time(); end - startin R. - If you’re satisfied with your analysis data and want to share it with others for their own analysis, use the
hise::uploadResultmethod to move the file into HISE. You can then find it through an advanced search. Use thehise::readFilesmethod to download it into an IDE.
Troubleshoot Package Installations
| Package | Error/Issue | Tips |
| sf | Linux dependency issues | Within an active Conda environment, enter the following command:conda install r-sf |
| sf | Linux dependency issues | In your terminal, enter the following command:apt-get -y update && apt-get install -y libudunits2-dev libgdal-dev libgeos-dev libproj-devThen try R installation. |
| Cairo | Linux dependency issues | In your terminal, enter the following command: Then try R installation. |
The accompanying table offers tips for troubleshooting Linux dependency issues with package installations. If you can't resolve the problem and and want to file a ticket, provide the notebook name, along with SessionInfo() and the instance name, in an email to immunology-support@allenimmunology.org.
Sometimes a particular step in your analysis, such as a clustering algorithm, is more computationally intensive than other steps. You can use a regular notebook to execute the nonintensive tasks interactively, and schedule an asynchronous job to execute the computationally demanding task. Once the latter is complete, you can see the results and derive further insights in the Notebook Jobs view. For instance, you might review the results of a clustering task executed as a notebook job, and then generate visualizations of those results.
Instructions
Schedule a notebook job
1. Log in to HISE, and navigate to RESEARCH > IDE instances.
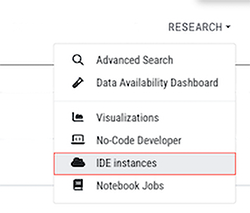
2. Choose any active instance, and click its title to open it.
3. In the side navigation, find the examples folder.
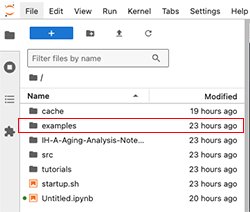
A. For an example that uses Seurat, open the following path:
/examples/R/Analysis/NotebookSchedulerSeurat.ipynb
B. For an example that uses h-sne, open the following path:
/examples/Python/Analysis/NotebookSchedulerDemo.ipynb
Check the status of a job
1. Log in to HISE, and navigate to RESEARCH > Notebook Jobs.
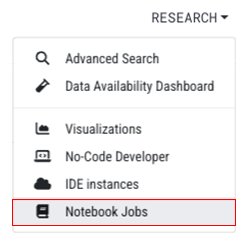
2. Find the job whose status you want to check. To select it, click the radio button at left.
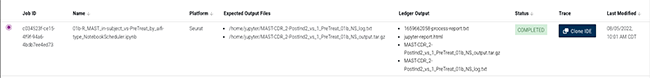
3. In the Ledger Output column, click process-report.txt.

4. In the Report links column at left, click Notebook Scheduler Report (txt).

A detailed report like this one appears.
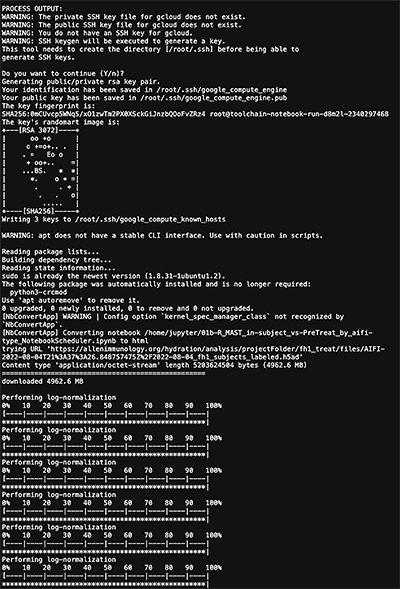
4. Alternatively or in addition, you can see the JSON file for the entire process. Beneath the Process name column, click Full JSON.

 Related Resources
Related Resources