Last modified 2026-01-20 |
Use HISE SDK Methods and Get Help in the IDE
 | Abbreviations Key | |||
| ad | AnnData (annotated data [Python/R package for storing spatial matrices]) | HTO | hashtag oligo | |
| ADT | antibody-derived tags | IDE | integrated development environment | |
| c | concatenate | QC | quality control | |
| HDF5 | hierarchical data format, version 5 | SDK | software development kit | |
| HISE | Human Immune System Explorer | stderr | standard error | |
| UMI | unique molecular identifier |
At a Glance
This document shows you how to get help on any given SDK method directly in the IDE. It also describes the SDK methods available in R and Python to read and download data to your instances, store intermediate files, schedule notebooks, manage IDE state, and save results, visualizations, or abstractions. If you have questions, contact Support.
|
You must preface each function call with the prefix Alternatively, to create an alias for |
Get Help in the IDE
For any given SDK method, you can get help without leaving the IDE. In this section, a cache_files() call in Python is used as the example throughout. For easy reference, the help methods are also shown in R, or you can use help.search("methodName") for syntax suggestions.
 | Python help(hp.method_name) | R help(methodName) |
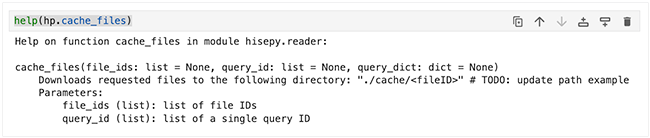
To display the function signature, list of parameters, class, and a brief description of the method as plain text, use the word help followed by the function name in parentheses (do not add a set of parentheses at the end of the function name):
help(hp.cache_files) |
 | Python hp.method_name? | R ?methodName or ?"methodName" |
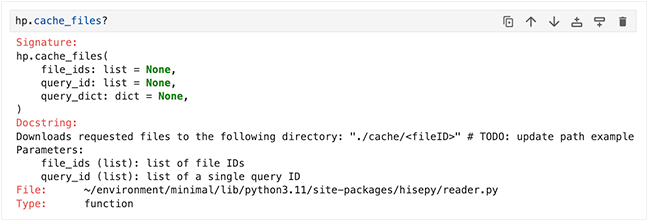
To display the signature and docstring (description) in a more readable format, which works well for complex signatures with lots of parameters, add a question mark at the end of the function name. This query also returns the file location and type:
hp.cache_files? |
 | Python hp.method_name?? | R hise::methodName |
If you want the whole enchilada—signature, docstring, file path, a verbose set of metadata, and the source code for the method, use a double question mark, like so:
hp.cache_files?? |

SDK Reader Methods
Following is a list of methods to read or download data to your IDE instance.
| Method Name | Description |
| hisepy.hise_file | Creates and returns a single hise_file object. |
| hisepy.cache_files | Downloads the specified files and stores them locally in a cache directory, without loading their contents into memory. |
| hisepy.query_files | Searches for files in HISE based on user-defined criteria, and returns metadata (but not file contents) for the matching files. |
| hisepy.cache_fileset | Downloads and caches metadata for a file set (a group of files) without loading file contents. |
| hisepy.list_filesets | Retrieves a list of available filesets, including their identifiers and basic metadata. |
| hisepy.get_file_descriptors | Retrieves file descriptors based on the user's query, as in this example:
|
| hisepy.read_files | Reads the contents of a list of file IDs into a hise_file object.Note: Only one parameter per function call is allowed. |
| hisepy.read_samples | Reads or searches the SampleStatus materialized view. Here's an example:
|
| hisepy.read_subjects | Reads or searches the
|
| Method Name | Description |
| cacheFiles() | Downloads the specified files and stores them locally in a cache directory, without loading their contents into memory. |
| getFileDescriptors() | Retrieves file descriptors based on the user's query. |
| readFiles() | Reads the contents of a list of file IDs into a hise_file object.Note: Only one parameter per function call is allowed. |
| readSamples() | Reads or searches the SampleStatus materialized view. |
| readSubjects() | Reads or searches the
|
SDK Upload Methods
The following methods help you upload results to the collaboration space.
| Method Name | Description |
| get_study_spaces | Returns a list of studies the user can access. |
| upload_files | Uploads files to a specified project or study. Can be used to save intermediate files that someone else can use as their starting point for further analysis. |
| save_visualization | Saves a Plotly object to a specified study. |
| load_visualization | Loads a Plotly visualization that was saved to a study. |
| save_dash_app | Given a Dash app consisting of an entry point named app.py and a list of supporting files, uploads and deploys that app to HISE as a visualization in the specified study. |
| save_static_image | Saves a static image (such as a PNG or JPEG file) to a specified study for documentation or sharing. |
| get_default_project | Retrieves the default project for the current user or session. |
| get_default_store | Retrieves the default data store for the current user or session. |
| get_files_for_query | Returns a list of files that match the specified query parameters within a study or project. |
| get_trace | Retrieves detailed trace or log information for a specific analysis, process, or workflow. |
| set_default_project | Sets the default project for the current user or session. |
| set_default_store | Sets the default data store for the current user or session. |
| save_custom_conda_environment | Saves a custom conda environment with additional metadata. |
| Method Name | Description |
| getStudySpace() | Returns a list of studies the user can access. |
| uploadFiles() | Uploads files to a specified project or study. Can be used to save intermediate files that someone else can use as their starting point for further analysis. |
| loadVisualization() | Loads a Plotly visualization that was saved to a study. Note: This function might be buggy. If so, contact Supportto file a ticket. |
| saveVisualization() | Saves a Plotly object to a specified study. Note: This function might be buggy. If so, contact Supportto file a ticket. |
| uploadFiles() | Uploads files to a specified project or study. |
Project Store SDK Methods
The following methods let you interact with the Project Store.
| Method Name | Description |
| delete_file_in_project_store | Deletes a file in the project store, provided it's not otherwise in use. |
| download_from_project_store | Downloads a given file to the user's IDE. |
| list_files_in_project_store | Returns a list of files present in a given project store. |
| list_project_stores | Lists all project stores the user can access. |
| promote_file_in_project_store | Marks a file in a project store to be promoted to the permanent store. After promotion, the file is managed in the permanent store and is no longer returned in a list_files_in_project_stores() call. |
| undo_delete_in_project_store | Undoes the file delete action, provided the call is executed within the file's retention period. |
| undo_promote_file_in_project_store | Undoes the promotion action, provided the file has not already been moved to the permanent store. After the undo call is executed, the file again becomes visible in a call to list_files_in_project_store(). |
| Method Name | Description |
| deleteFileInProjectStore() | Deletes a file in the project store, so long as it is not otherwise in use. |
| downloadFileFromProjectStore() | Downloads a given file onto a user's IDE. |
| listFilesInProjectStores() | Returns information about what files are present in a given project store. |
| listProjectStores() | Lists all project stores a user has access to. |
| promoteFileInProjectStore() | After promotion, the file is managed in the permanent store and is no longer returned in a listFilesInProjectStore() call. |
| undoDeleteFileInProjectStore() | Undoes the file delete action, so long as it is within the file's retention period. |
| undoPromoteFileInProjectStore() | Undoes the promotion action, so long as the file has not already been moved to the permanent store. The file will once again be visible via listFilesInProjectStore(). |
SDK Private Folder Methods
The following methods let you interact with private folders.
| Method Name | Description |
| create_private_folder | Creates a new private folder. |
| delete_file_in_private_folder | Deletes a file from a private folder. |
| delete_private_folder | Deletes an existing private folder. |
| download_from_private_folder | Downloads a file from a private folder to your local working directory. |
| find_private_folder_of_file | Returns the name or identifier of the private folder containing the specified file. |
| list_files_in_all_private_folders | Returns a data.frame of all private folders and files that are within each. |
| list_files_in_private_folder | Lists files inside a given private folder. |
| move_file_in_private_folder | Moves a file between private folders. |
| rename_file_in_private_folder | Renames a file in a private folder. |
| upload_file_to_private_folder | Uploads a file to a private folder. |
| Method Name | Description |
| createPrivateFolder() | Creates a new private folder. |
| deleteFileInPrivateFolder() | Deletes a file from a private folder. |
| deletePrivateFolder() | Deletes an existing private folder. |
| downloadFromPrivateFolder() | Downloads a file from a private folder to your local working directory. |
| listFilesInAllPrivateFolders() | Lists all folders that belong to this user and the files inside them. |
| listFilesInPrivateFolder() | Lists files inside a given private folder. |
| MoveFileInPrivateFolder() | Moves a file between private folders. |
| renameFileInPrivateFolder() | Renames a file in a private folder. |
| uploadFileToPrivateFolder() | Uploads a file to a private folder. |
IDE Management SDK Methods
The following method stops an IDE instance.
| Method Name | Description |
| stop_ide | Stops the active instance from which this method is invoked. |
| Method Name | Description |
| stopIDE() | Stops the active instance from which this method is invoked. |
SDK Lookup Methods
Lookup methods are used to find appropriate field names for querying, and to look up the unique values for each field name.
| Method Name | Description |
| lookup_queryable_fields | Returns a list of fields used for creating queries. |
| lookup_unique_entries | Returns unique values for a given field. |
| Method Name | Description |
| lookupQueryableFields() | Returns a list of fields used for creating queries. |
| lookupUniqueEntries() | Returns unique values for a given field. |
SDK Abstraction Methods
Below are a few SDK methods to help you work with abstractions.
| Method Name | Description |
| get_result_files | Retrieves a list of result files generated by an analysis or workflow. |
| result_filetype_to_guid | Returns the GUID associated with the specified result file type. |
| save_abstraction | Saves a visualization template to the IDE user's account. |
SDK Utilities
Following is a brief list of utility methods.
| Method Name | Description |
| _dict_to_df | Converts a data dictionary into a pandas DataFrame for further analysis or processing. |
| hise_file_to_df | Converts the contents of a hise_file object into a pandas DataFrame. |
| sample_to_df | Converts sample-level metadata or data into a pandas DataFrame. |
| subject_to_df | Converts subject-level metadata or data into a pandas DataFrame. |
AI/ML Training Run SDKs
Following is a brief list of methods associated with AI/ML training runs.
| Method Name | Description |
| start_training_run | Launches a remote, resource-configurable training job for an AI/ML model. |
| review_training_job_run | Lets users review and then approve or reject an AI/ML training job. |
| conda_env_builds | Returns True if conda env can build successfully, False otherwise. |
H5 Weaver SDK Methods
The following R functions are for reading, rearranging, and writing 10x-compatible HDF5 files.
| Method Name | Description |
| add_cell_ids | Adds cell IDs to an h5_list object. |
| add_h5_list_adt_results | Adds ADT results to an h5_list. |
| add_h5_list_hash_results | Adds cell hashing results to an h5_list object. |
| add_h5_transposed_matrix | Adds a transposed version of a matrix to an .h5 file. |
| add_well_metadata | Adds well metadata. |
| append_ext_h5_character | Appends character values to an existing HDF5 string dataset. |
| cat_h5_list | Concatenates two h5_list objects. |
| choose_chunk_size | Select a reasonable chunk size for an HDF5 dataset object. |
| choose_integer_bits | Decides how many bits to use to store integer values in an HDF5 dataset object. |
| convert_char_na | Converts NA character entries to actual NAs. |
| convert_char_na_recursive | Converts all NA character values to actual NAs recursively. |
| create_ext_h5_character | Creates an extensible character HDF5 dataset. |
| create_ext_h5_float | Creates an extensible float HDF5 dataset. |
| create_ext_h5_uint | Creates an extensible unsigned integer HDF5 dataset. |
| get_list_path | Retrieves an object from a list using path-style targeting. |
| h5_attr_list | Generates a set of attributes based on 10x Genomics defaults. |
| h5_list_add_dgCMatrix | Adds a sparse matrix to an h5_list containing 10x Genomics data. |
| h5_list_add_mita_umis | Adds mitochondrial gene UMI counts to an h5_list object. |
| h5_list_cell_metadata | Extracts a data.frame of cell metadata from an h5_list object. |
| h5_list_convert_from_dgCMatrix | Converts the matrix in an h5_list from a sparse matrix back to its original structure. |
| h5_list_convert_to_dgCMatrix | Converts the matrix in an h5_list from 10x Genomics data to a sparse matrix. |
| h5_list_transpose | Transposes an h5_list object. |
| h5dims | Gets dimensions of an object in an HDF5 file. |
| h5dump | Dumps all objects from an HDF5 file to a list. |
| h5exists | Checks if an object exists in an HDF5 file. |
| h5ls | Lists objects in an HDF5 file. |
| qc_aligned_barplot | Generates a baseline-aligned barplot for two categorical metrics. |
| qc_cutoff_barplot | Generates a QC barplot for a metric at multiple cutoffs. |
| qc_flip | Flips the orientation of a data.frame and returns as a list. |
| qc_frac_hist_plot | Generates a QC histogram plot for a single fractional metric. |
| qc_hist_plot | Generates a QC histogram plot for a single metric. |
| qc_stacked_barplot | Generates a stacked barplot for two categorical metrics. |
| qc_table | Renders a custom-formatted table for QC metrics. |
| qc_violin_plot | Generates a QC violin plot for a metric, grouped by a categorical metadata column. |
| read_h5_cell_meta | Reads .h5 cell metadata. |
| read_h5_dgCMatrix | Reads the /matrix from an .h5 file as a sparse matrix. |
| read_h5_feature_meta | Reads .h5ad feature metadata. |
| read_tenx_metrics | Reads and corrects formatting of a 10x metics_summary.csv file. |
| reduce_h5_list | Reduces a list-of-lists of h5_list objects to a single, concatenated object. |
| set_list_path | Sets a list value using path-style targeting. |
| split_h5_list_by_hash | Splits a 10x HDF5 file based on HTOparser results. |
| stm | Writes a stderr message with a leading data/time stamp. |
| strip_1d_array_recursive | Recursively traverses a list and converts into vectors any one-dimensional array elements it contains. |
| subset_h5_list_by_observations | Subsets an h5_list object based on a set of observations. |
| varibow | Generates a rainbow palette with variations in saturation and value. |
| write_10x_h5_container | Generates an empty HDF5 container to store RNA-seq count data in 10x format. |
| write_h5_list | Writes an h5_list, as created by rhddf5::h5dump(), to an .h5 file. |
List of All SDK Modules and Methods
Sometimes it's helpful to have a list of all modules and methods in alphabetical order. This is that list.
abstraction auth cache_files cache_fileset clear_notebook_job conda_pack conda_env_builds delete_file_in_private_folder delete_file_in_project_store delete_private_folder download_from_private_folder download_from_project_folder download_from_project_store find_private_folder_of_file get_default_project get_default_store get_file_descriptors get_files_for_query get_memory_usage get_notebook_job get_result_files get_study_spaces get_trace hise_file hise_file_to_df list_files_in_all_private_folders list_files_in_private_folder list_files_in_project_folder list_files_in_project_store list_filesets list_project_folders list_project_stores load_visualization lookup lookup_queryable_fields lookup_unique_entries | move_file_in_private_folder notebook_job private_folders project_folder project_store promote_file_in_project_store query_files read_files read_samples read_subjects rename_file_in_private_folder result_filetype_to_guid retry_ide_commit review_training_job_run sample_to_df save_abstraction save_custom_conda_environment save_dash_app save_static_image save_visualization schedule_notebook set_default_project set_default_store set_memory_limit start_training_run stop_ide subject_to_df training_job undo_delete_in_project_store undo_promote_in_project_store update_sdk_version upload upload_file_to_private_folder upload_files version |
assembleQuery buildDataFrame buildDescriptors buildSparseMatrix cacheCytometryFile cacheCytometryResult cacheFileSets cacheFiles clearDownloadCache clearNotebookJob condaEnvBuilds convertFileToCytoResponse convertFilelistToCytoResponse convertObjectToCytoResponse convertObjectToMFIResponse deleteFileInPrivateFolder deleteFileInProjectStore downloadFileFromPrivateFolder downloadFileFromProjectFolder downloadFileFromProjectStore downloadJobOutput findPrivateFolderOfFile getDefaultProject getDefaultStore getFileDescriptors getMemoryUsage getNotebookJob getParam getStudySpaces getTrace getUniqueLabResults listFileSets listFilesInAllPrivateFolders listFilesInAllProjectFolders listFilesInAllProjectStores listFilesInPrivateFolder listFilesInProjectFolders listFilesInProjectStores listParams listProjectFolders listProjectStores listProjects | loadGatingSet loadParams loadVisualization lookupQueryableFields lookupUniqueEntries moveFileInPrivateFolder prepareFile promoteFileInProjectStore readBridgingControlForCytoMFIFile readBridgingControlForCytometryFile readCytoMFIFile readCytoMFIResult readCytometryFile readCytometryResult readFiles readFiles_v1 readQuery readSamples readSubjects renameFileInPrivateFolder runQuery saveCustomCondaEnvironment saveParams saveStaticImage saveVisualization scheduleNotebook setCurrentProject setDefaultProject setDefaultStore setFileMetadataInProjectFolder setFileMetadataInProjectStore setMemoryLimit setParam setProjects stopIDE undoDeleteFileInProjectStore undoPromoteFileInProjectStore updateSDKVersion uploadFileToPrivateFolder uploadFiles uploadFiles_v1 viewParams |
 HISE SDK User Documentation
HISE SDK User Documentation Related Resources
Related Resources